News Release | Deep Docking software is fast-tracking research into effective medications designed specifically to treat the COVID-19 causing virus SARS-CoV-2.
Vancouver, B.C. – Researchers at the Vancouver Prostate Centre (VPC) have engineered the fastest, artificial intelligence (AI)-enabled technology of its kind to identify novel medications for the SARS-CoV-2 virus. The team’s published study explains how they were able to supercharge the computational screening for potential antiviral compounds more than 100-fold, opening the door to the rapid development of medications to treat infected individuals.
“While there are several vaccines on the market, there is currently no medication to treat COVID-19. Our Deep Docking technology enables easier and faster identification of prospective COVID-19 medications at the molecular level,” says lead investigator, Dr. Artem Cherkasov, a senior scientist with the Vancouver Coastal Health Research Institute (VCHRI) and professor in the Department of Urologic Sciences at the University of British Columbia (UBC).
“Only when both the vaccine and a medication are available can we most efficiently manage COVID-19. Right now we are working with one arm, and we need two,” says Cherkasov.

Deep Docking is a computer-aided drug discovery protocol. Designed by Cherkasov, this AI-enabled approach can screen hundreds of billions of molecular structures to see how they fit into the viral target protein—an approach also known as molecular docking. Cherkasov’s latest research—published in Chemical Science, the Royal Society of Chemistry’s flagship journal—was able to perform molecular docking at a speed never before achieved.
“It took us 19 days to complete the Deep Docking of 40 billion compounds. This formerly would have taken 10 years with the fastest available technology,” says Cherkasov.
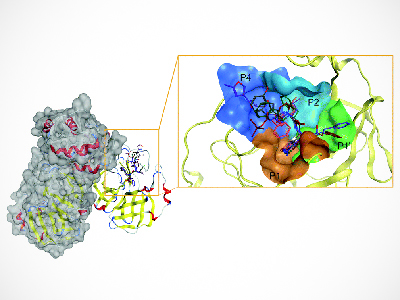
The study team used Deep Docking to target SARS-CoV-2’s main protease (Mpro)—an enzyme that is important for its viral replication. They then docked over 40 billion virtual small molecules into the Mpro, funneling down to a handful that were able to effectively inhibit enzymatic Mpro’s activity. Five docking programs were employed as cross-references.
“Mpro can be thought of as a lock. The small molecules we screened are keys we tested in the lock to identify which ones could open Mpro and disable it,” explains Dr. Larry Goldenberg, director of development and supportive care at the VPC and a professor in the Department of Urologic Sciences at UBC.
“This is a great example of how the research team was able to turn on a dime when COVID-19 hit, and pivot our expertise in cancer drug design to COVID-19 drug design,” says Goldenberg.
If a medication for COVID-19 was discovered, it could be used to help infected individuals recover faster, cutting down on hospital admissions and improving patient outcomes, says Cherkasov, who has been investigating the application of AI in drug discovery since the early 2000s. He adds that it could also be used as a prophylactic to prevent infection with the virus, which has already claimed the lives of over five million people globally.
All the data collected by the team will be publicly accessible through Github and the Democratizing Drug Discovery with Deep Docking (D5) platform being developed by Cherkasov, which “will give scientists from around the world rapid access to billions of molecules that they can use for their research.”
The ingredients of any COVID-19 medication developed by the team will also be shared with the world for free. “We are doing this because we want everyone to have access to this information, to save the world in a way,” says Cherkasov.
This research was made possible with support from Telus, the 625 Powell Street Foundation, Teck Resources Limited, NVIDIA, Dell Technologies HPC and AI Innovation Lab, R & J Stern Family Foundation, VGH & UBC Hospital Foundation and several private donors.


